library(tidyverse)
library(palmerpenguins)
theme_set(theme_bw())
theme_update(axis.title = element_text(size = 24))
tuesdata <- tidytuesdayR::tt_load(2020, week = 28)#>
#> Downloading file 1 of 1: `coffee_ratings.csv`Statistical Inference
Power Analysis
Resampling Techniques
Permutation Tests
a1 <- data.frame(
x = c(2.5,3.5),
y = c(41, 41),
label = c("Yes", "No")
)
a2 <- data.frame(
x = c(1.9),
y = c(25, 35),
label = c("False", "True")
)
a3 <- tibble(x = c(1.7,3),
y = c(30, 43),
label = c("H0", "Reject H0"))
yay <- tibble(x = c(2.5, 3.5),
y = c(25, 35),
label = c("Yay!", "Yay!"))
type1 <- tibble(x = c(2.5, 3.5),
y = c(36.5, 26.5),
label = c("Type I Error", "Type II Error"))
type2 <- tibble(x = c(3.5, 2.5),
y = c(24.5, 34.5),
label = c("beta", "alpha"))
# basic graph
p <- ggplot() + theme_void()
# Add rectangles
p + annotate("rect",
xmin=c(2,3), xmax=c(3,4),
ymin=c(20,20), ymax=c(30,30),
alpha=0.2, color="green", fill="green") +
annotate("rect",
xmin=c(2,3), xmax=c(3,4),
ymin=c(30,30), ymax=c(40,40),
alpha=0.2, color="red", fill="red") +
annotate("rect",
xmin=c(3,2), xmax=c(4,3),
ymin=c(30, 20), ymax=c(40, 30),
alpha=0.8, color="royalblue", fill="royalblue") +
geom_text(data=a1, aes(x=x, y=y, label=label),
size=10 , fontface="bold" ) +
geom_text(data=a3, aes(x=x, y=y, label=label),
size=10 , fontface="bold.italic" ) +
geom_text(data=a2, aes(x=x, y=y, label=label),
size=10, angle = 90, fontface="bold" ) +
geom_text(data=yay, aes(x=x, y=y, label=label),
size=10, fontface="bold.italic" ) +
geom_text(data=type1, aes(x=x, y=y, label=label),
size=10, fontface="bold") +
geom_text(data=type2, aes(x=x, y=y, label=label),
size=10, fontface="bold", parse = T ) x <- seq(-4, 4, length.out = 1000)
xx <- seq(-1, 7, length.out = 1000)
dt_one<-function(x){
y <- dnorm(x)
y[x < qnorm(.94)] <-NA
return(y)
}
dt_two<-function(x){
y <- dnorm(x, mean = 3)
y[x > 1.645] <-NA
return(y)
}
df1 <- tibble(x = x, y = dnorm(x))
df2 <- tibble(x = xx, y = dnorm(xx, mean = 3))
a1 <- tibble(x = c(0,3), y = c(0.43, 0.43), label = c("H0", "H1"))
a2 <- tibble(x = c(2.05,1.25), y = c(0.02, 0.02), label = c(paste("alpha"), "beta"))
df1 |>
ggplot(aes(x, y)) +
stat_function(fun = dt_one, geom = "area", fill = "red") +
geom_line() +
geom_line(aes(x, y), df2) +
stat_function(fun = dt_two, geom = "area", fill = "green") +
geom_vline(aes(xintercept = 1.645), lwd = 1.5) +
geom_text(data=a1, aes(x=x,y=y, label=label),
size=14 , fontface="bold.italic" ) +
geom_text(data=a2, aes(x=x,y=y, label=label),
size=10 , fontface="bold.italic", parse = T) +
theme_bw()Power is the probability of rejecting \(H_0\) given that \(H_1\).
\[ Power = 1-\beta \]
We want to ensure we have a high power and low type I error rate (\(\alpha\)).
As practitioners, we control \(\alpha\). Set it before we conduct a study. Usually set at 0.05.
We cannot control power because it is dependent by effect size, \(\alpha\), and sample size.
The pwr package in R will conduct power and sample size for several common statistical tests. For more information, you can check this vignette and their github.
x <- seq(-4, 4, length.out = 1000)
xx <- seq(-1, 7, length.out = 1000)
dt_one<-function(x){
y <- dnorm(x)
y[x < qnorm(.94)] <-NA
return(y)
}
dt_two<-function(x){
y <- dnorm(x, mean = 3)
y[x > 1.645] <-NA
return(y)
}
df1 <- tibble(x = x, y = dnorm(x))
df2 <- tibble(x = xx, y = dnorm(xx, mean = 3))
a1 <- tibble(x = c(0,3), y = c(0.43, 0.43), label = c("H0", "H1"))
a2 <- tibble(x = c(2.05,1.25), y = c(0.02, 0.02), label = c(paste("alpha"), "beta"))
df1 |>
ggplot(aes(x, y)) +
stat_function(fun = dt_one, geom = "area", fill = "red") +
geom_line() +
geom_line(aes(x, y), df2) +
stat_function(fun = dt_two, geom = "area", fill = "green") +
geom_vline(aes(xintercept = 1.645), lwd = 1.5) +
geom_text(data=a1, aes(x=x,y=y, label=label),
size=14 , fontface="bold.italic" ) +
geom_text(data=a2, aes(x=x,y=y, label=label),
size=10 , fontface="bold.italic", parse = T) +
theme_bw()Given a Null and Alternative hypothesis, one can determine how often a you will reject the null hypothesis from a high number of simulated data.
This is done by simulating from a hypothesized alternative distribution, conducting a hypothesis test given that the null hypothesis is true, and determine how often do you reject the null hypothesis.
\[ Power = \frac{\#\ Rejected}{\#\ Simulated\ Data} \]
Use a Monte Carlo Hypothesis Test for \(H_0: \mu = 6\) and \(H_a: \mu \neq 6\). Simulate fake data from 24 RV’s from \(N(3, 9)\). Testing how the distribution will look like assuming that the null hypothesis is true.
Answer:
tstat <- (mean(x) - 6)/(sd(x)/sqrt(24))
t_stat_sim <- function(i){
x <- rnorm(24, 6, 3)
tt <- (mean(x) - 6)/(sd(x)/sqrt(24))
return(tt)
}
t_dist <- sapply(1:100000, t_stat_sim)
t_dist |> tibble(x = _) |>
ggplot(aes(x, y = ..density..)) +
geom_histogram() +
geom_vline(aes(xintercept = tstat), col = "red")
sum(abs(tstat) < abs(t_dist))
(sum(abs(tstat) < abs(t_dist)) + 1) / (length(t_dist) + 1)alpha <- 0.05
cv <- qt(1-alpha/2, 23)
t_stat_sim <- function(i, mu){
x <- rnorm(24, mu, 3)
tt <- (mean(x) - 6)/(sd(x)/sqrt(24))
return(cv < abs(tt))
}
mus <- 0:12
powers <- c()
for(i in 1:13){
t_dist <- sapply(1:100000, t_stat_sim, mu = mus[i])
powers <- c(powers, mean(t_dist))
}
tibble(x = mus, y = powers) |>
ggplot(aes(x, y)) +
geom_line() +
ylab("Power") +
xlab("Alternative")Power Analysis
Resampling Techniques
Permutation Tests
Resampling techniques involve methods that require to sample from the data, instead of the parametric model. Common Methods:
Given a data set, we sample an observation to a new data set. The sampled observation can be resampled again to the new data set.
Given a data set, we sample an observation to a new data set. The sampled observation cannot be resampled again to the new data set.
Power Analysis
Resampling Techniques
Permutation Tests
Permutation tests conducts a statistical test by constructing the null distribution by rearranging the data points in a sample.
Null hypothesis states that the rearrangements of the data points are random.
Alternative hypothesis states that the rearrangement of the data points aren’t random.
\[ F_x = F_y \]
\[ F_x \neq F_y \]
Suppose \(\{X_i, Y_i\}^n_{i=1}\) is an observed permutation, \(X = \{X_1, \ldots, X_n\}\), \(Y = \{Y_1, \ldots, Y_n\}\).
The Probability of any permuation is \(1/n!\).
Therefore, for a statistic \(T(X,Y)\), a sampling distribution can be constructed by all the different permutations.
A hypothesis test can be conducted by observing the proportion of more extreme values of the sample statistic.
Constructing the distribution for the permutations can be challenging if the number of permutations is high! If \(n=100\), the number of permutations is \(100!\):
Therefore, simulation techniques are needed to approximate the p-value.
By randomly drawing from the sample, we can approximate the p-value.
\[ p = \frac{m +1}{N + 1} \]
penguins |> ggplot(aes(x = species, y = body_mass_g)) +
labs(x = "Species", y = "Body Mass") +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(aes(species, shuffle(body_mass_g))) +
geom_jitter(col = "red") We want to determine if body mass of penguins are different for different species.
#> Analysis of Variance Table
#>
#> Response: body_mass_g
#> Df Sum Sq Mean Sq F value Pr(>F)
#> species 2 145190219 72595110 341.89 < 2.2e-16 ***
#> Residuals 330 70069447 212332
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1f_stat <- penguins |>
aov(body_mass_g ~ species, data = _) |>
anova() |>
_$`F value`[1]
f_sim <- function(i){
ff <- penguins |>
aov(shuffle(body_mass_g) ~ species, data = _) |>
anova() |>
_$`F value`[1]
return(ff)
}
f_dist <- replicate(10000, f_sim(1))
tibble(x= f_dist) |>
ggplot(aes(x, y = ..density..)) +
geom_histogram() +
geom_vline(xintercept = f_stat)#> [1] 9.999e-05Is there a linear relationship between flavor and aroma in coffee drinks from the coffee_aroma data set.
coffee_aroma |> ggplot() +
geom_smooth(mapping = aes(aroma, flavor), method = "lm", se = F, col = "red") +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) +
geom_smooth(mapping = aes(aroma, shuffle(flavor)), method = "lm", se = F) f_stat <- coffee_aroma |>
lm(flavor ~ aroma, data = _) |>
_$`coefficients`[2]
f_sim <- function(i){
ff <- coffee_aroma |>
lm(shuffle(flavor) ~ aroma, data = _) |>
_$`coefficients`[2]
return(ff)
}
f_dist <- replicate(10000, f_sim(1))
tibble(x= f_dist) |>
ggplot(aes(x, y = ..density..)) +
geom_histogram() +
geom_vline(xintercept = f_stat)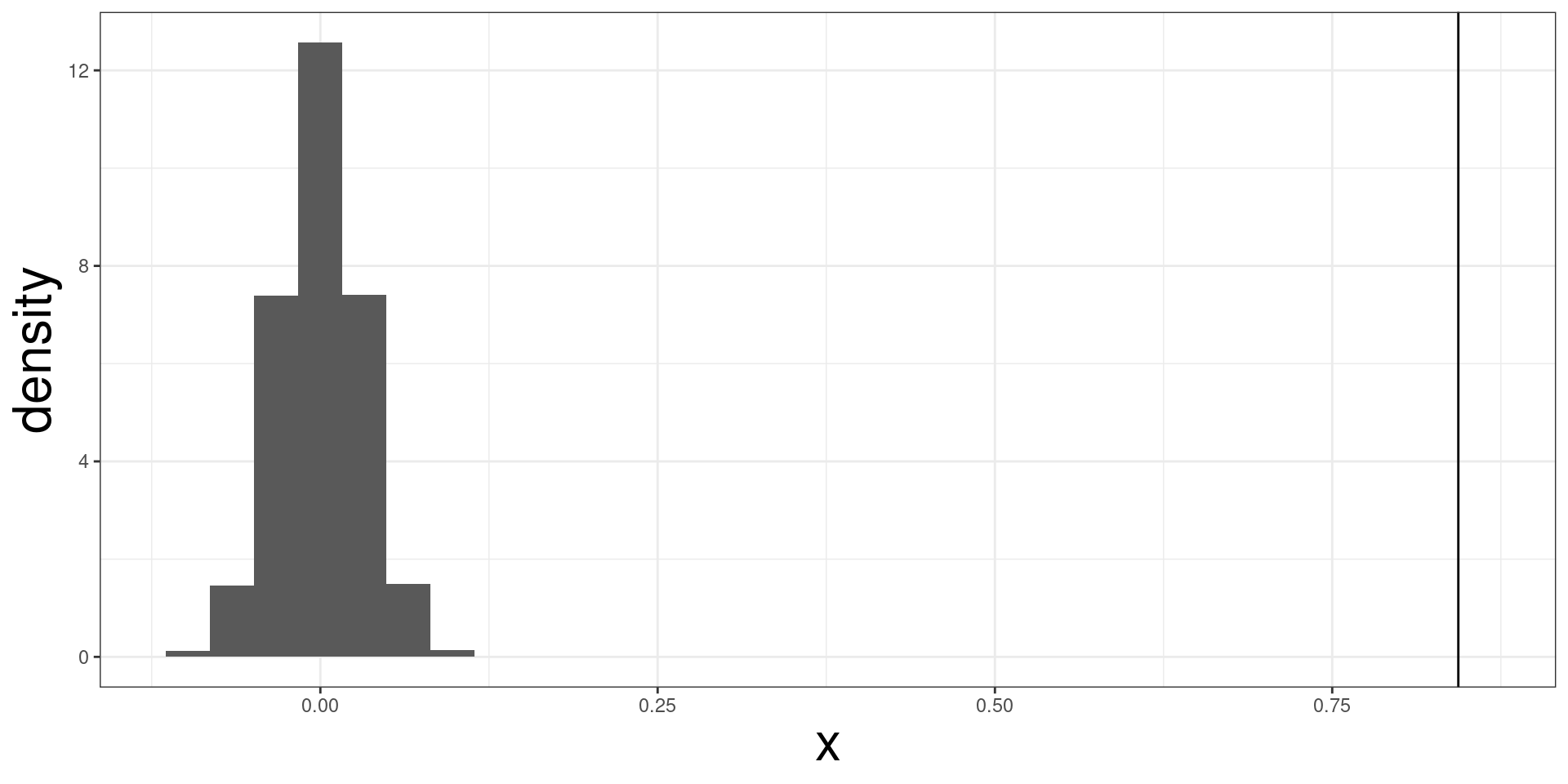
#> [1] 9.999e-05